Welcome to PELE Web Server
This website is free and open for non-commercial usage, with a limit of 16 CPUs per simulation. Commercial users and those users needing full support and no limit in the number of CPUs, please contact Nostrum Biodiscovery (http://nostrumbiodiscovery.com).
You have to be a registered user (see register page) in order to use the server. Then, you will have to be logged in to submit jobs (see login page).
About PELE
Using technological advances in protein structure prediction, we have developed PELE (an acronym for Protein Energy Landscape Exploration), a novel method to perform protein energy landscape explorations. PELE combines a Monte Carlo stochastic approach with protein structure prediction algorithms and is capable of accurately reproducing long time scale processes in only few hours of CPU.
PELE combines a Monte Carlo stochastic approach with protein structure prediction. The Figure below shows the heuristic algorithm for the landscape exploration method which is based on three main steps: an initial perturbation, a side chain sampling and a final minimization.
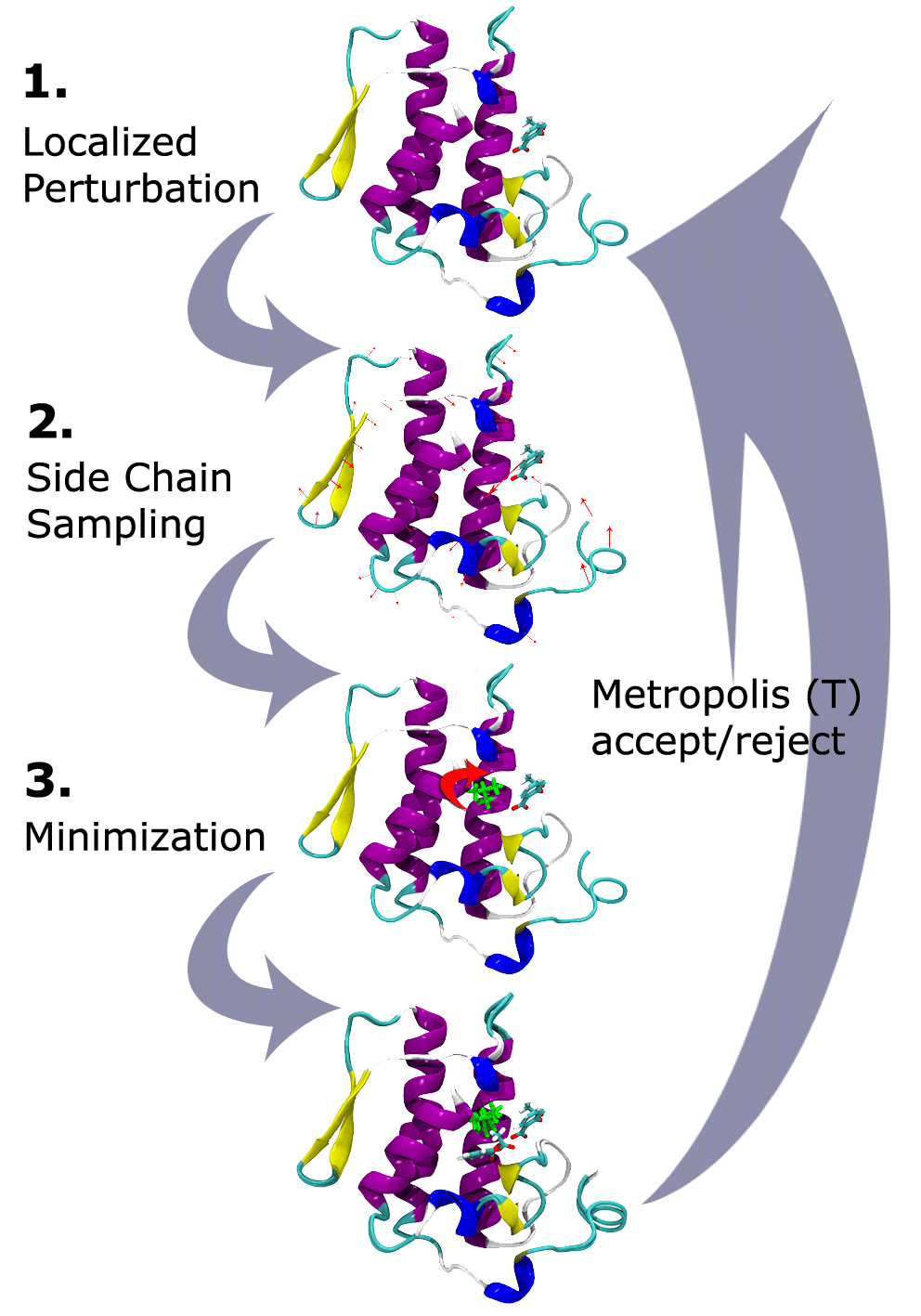
Localized perturbation. After an energy calculation for the initial structure, the procedure begins with the generation of a perturbation in the system. In studies of ligand diffusion or ligand induced fit, the perturbation starts with a random translation and rotation of the ligand. The perturbation might include the backbone of the protein (or the backbone surrounding the ligand) by performing a minimization (all atom) where the alpha carbons are driven to a new position resulting from a small displacement in a low frequency anisotropic normal mode (ANM). Such a procedure aims to describe to the global motion of the protein.
Side chain sampling. The algorithm proceeds by placing all side chains local to the ligand with a rotamer library side chain optimization at a rotamer resolution of 10°. The side chain algorithm uses steric filtering and a clustering method to reduce the number of rotamers to be minimized (side chains minimization only).
Minimization. The last step involves the minimization of a user-defined region (typically we include the full protein) with a Truncated Newton minimizer.
These three steps compose a move that is accepted (defining a new minima) or rejected based on a Metropolis criterion. The procedure has been parallelized using the MPI communications protocol, with the option of interchanging coordinates between different trajectories. Whenever any trajectory is significantly further along a given reaction coordinate than any of the other trajectories, the trailing trajectory is abandoned and restarted from the position of the leading trajectory. This allows an efficient sampling of the configurational space towards one defined objective: entrance/escape of the ligand, optimization of the protein-ligand binding energy, etc.
Some example studies of PELE
- Ligand migration in truncated hemoglobin II [J. Biol. Chem., 284:3106-3116 (2009)]
- Induced fit docking [J. Comp. Chem., 31:1224-35 (2010)]
- Ligand binding in Aryl Alcohol Oxidase [Biochemical Journal, 436:341-350 (2011), J.Biol. Chem., 286:41105-41114 (2011)]
- Ligand migration in Human Hemoglobin [Biophysical Journal, 102:887-896 (2012)]
- Ligand binding in mTOR [Biochemical Pharmacology, 83:489-496 (2012)]
- Protein conformational sampling [J. Chem. Theo. Comput., 8:959-965 (2012)]
PELE original publications
- PELE: Protein energy landscape exploration. A novel Monte Carlo based technique [J. Chem. Theo. Comput., 1(6):1304-1311 (2005)]
- PELE web server: atomistic study of biomolecular systems at your fingertips [Nucleic Acids Research, 41(W1):W322-W328 (2013)]


